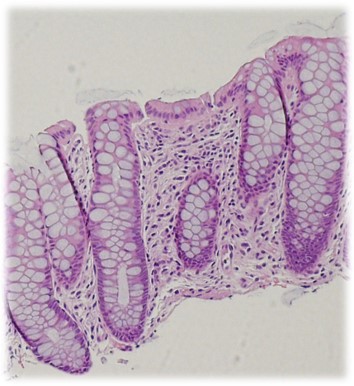
BarcUVa-Seq is a public-health genomics study of gene expression in the human colon. The principle aim is to characterize regulation of transcription in superficial epithelial cells across different colon subsites. We measure effects of variables such as genotype, sex, colon subsite, lifestyle, and diet on the composition of healthy colon transcriptomes.
BarcUVa-Seq is a population-based, cross-sectional study of 485 adult volunteers found to have endoscopically healthy (i.e normal, without polyps or other lesions) colon. Transcriptomic profiles were measured with bulk RNA sequencing of total RNA from superficial mucosal biopsies collected at different subsites across the colon during colonoscopy. Genotypes were measured with the OncoArray 500K BeadChip using DNA from peripheral blood samples.
We find transcriptional differences between anatomic subsites in the colon. We also find genotype-transcription level associations at thousands of variants across the genome. Many of the transcription-associated variants co-localize with disease risk loci identified in genome-wide association studies of various diseases. The BarcUVa-Seq dataset is a valuable resource for researchers interested in colon-related phenotypes and other traits.
Papers
Dampier, C. H. et al. Oncogenic Features in Histologically Normal Mucosa: Novel Insights Into Field Effect From a Mega-Analysis of Colorectal Transcriptomes. Clin. Transl. Gastroenterol. 11, e00210 (2020).
Diez-Obrero, V. et al. Genetic effects on transcriptome profiles in colon epithelium provide functional insights for genetic risk loci. Cell Mol Gastroenterol Hepatol (2021) doi:10.1016/j.jcmgh.2021.02.003.

Graham Casey UVA -PI

Victor Moreno ICO -PI

Virginia Díez-Obrero ICO

Chris H Dampier UVA

Matthew Devall UVA

Sarah J Plummer UVA

Ferran Moratalla-Navarro ICO

Elisabet Guinó ICO

Mireia Obón-Santacana ICO

Anna Díez-Villanueva ICO

Gemma Ibáñez-Sanz ICO

Robert Carreras-Torres ICO
This is a joint project of
In addition to ICO and UVA, this project has received support from: